
Research
Unraveling the Molecular Secrets of Ion Channel and Prenyltransferase Regulation
Enzymes are the driving force behind every cellular function, acting as biological catalysts that accelerate chemical reactions with incredible precision. Most enzymes are proteins, and their ability to speed up reactions depends on their intricate structure, dynamic movements, and allosteric regulation.
Our research is dedicated to understanding how protein structure drives their translates to their cellular roles.
With ion channels and prenyltransferases playing key roles in many diseases, our goal is to provide new molecular insights that could pave the way for targeted therapeutic approaches in the future. Our current research interests include:
-
Understanding how the specific molecular architectures are funneled into specialized activity(ies).
-
Illuminating the molecular mechanism of ion channels and prenyltransferase dysfunction in human diseases, with future prospects of enabling the development of targeted therapeutic strategies.
To do so, we use a wide array of approaches:
Structural biology

Visualize protein structures at the atomic level
Protein biochemistry
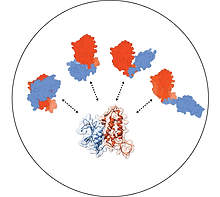
Monitor protein activity in predefined environments
Electrophysiology
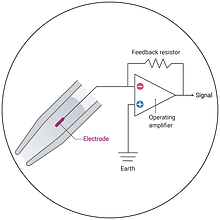
Study protein function at an unparalleled temporal resolution
Current research projects
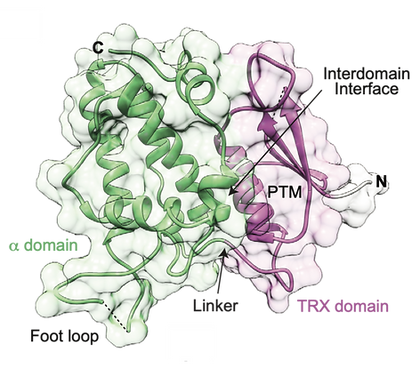
The cryptic family of Chloride Intracellular Channels (CLICs) consists of unique proteins, that were suggested to adopt both soluble and membrane-associated forms. Following this unusual metamorphic change, CLICs were shown to incorporate into membranes and mediate ion conduction in vitro, suggesting multimerization upon membrane insertion. Recently, we provided mechanistic insights into the metamorphic nature of human CLIC5 and mouse CLIC6. Using X-ray crystallography, SAXS, and mass spectrometry analyses we demonstrated that members of the CLIC family are inherently flexible. Additionally, we showed that oxidative conditions result in the dioxidation of a specific conserved cysteine residue, resulting in increased exposure of cryptic hydrophobic surface and membrane interaction. Currently, we uncovered a novel activity of this curious protein family!
The molecular basis for chloride intracellular channels unique functional specialization
Studies of cancer-related mechanisms involving aberant prenyltransferase activity
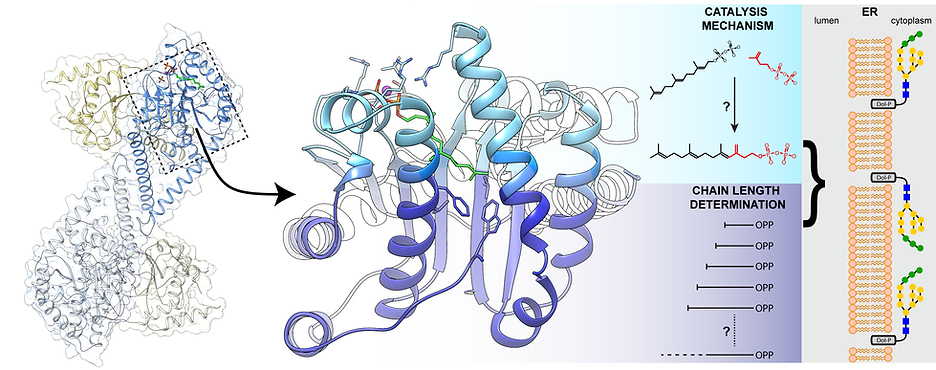
Cancer cells acquire hallmark properties that arm them with abnormally enhanced proliferative potential and reduced dependence on environmental cues, leading to tumor growth and metastasis. These hallmarks are obtained in part by extensively rewiring metabolic pathways essential for cell survival. Rewired post-translational protein modifications support the malignant characteristics of cancer cells. Thus, inhibition of key enzymes in this pathway represents a novel yet under-explored therapeutic venue for anti-cancer drug design. Our group has recently established a multidisciplinary approach to elucidate the structural and functional properties of prenyltransferases, key enzymes crucial for various post-translational modification pathways, which may serve as viable and novel targets for cancer treatment. In this project, using cutting-edge multi-disciplinary molecular approaches, we aim to elucidate the structural mechanisms governing prenyltransferases’ function. Furthermore, with our overarching goal of developing novel therapeutic approaches for cancer, we currently establish and utilize high-throughput platforms for the identification of agents that target and inhibit members of this enzyme superfamily.
Determining the interplay between KCNH channels intracellular domains and cancerous proliferation
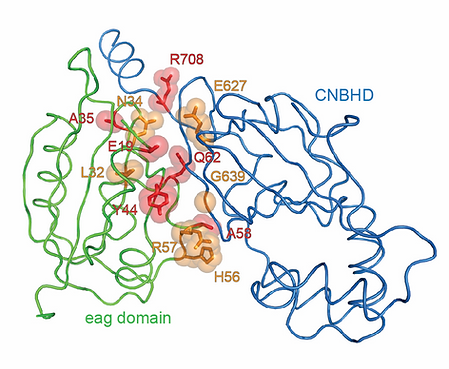
The KCNH voltage-dependent potassium channels are important regulators of cellular excitability and have key roles in diseases such as cardiac long QT syndrome type 2 (LQT2), epilepsy, schizophrenia, and cancer. The intracellular domains of KCNH channels are structurally distinct from other voltage-gated channels. The amino-terminal region contains an eag domain, which is composed of a Per-Arnt-Sim (PAS) domain and a PAS-cap domain, whereas the carboxy-terminal region contains a cyclic nucleotide-binding homology domain (CNBHD), which is connected to the pore through a C-linker domain. By mapping disease-associated mutations onto recently solved structures of KCNH family members, we aim to gain unique insights into the physiological and pathophysiological mechanisms of KCNH channels. Recently, we solved the first high-resolution structure of the CNBHD of the human KCNH2 (hERG; Ben-Bassat et al., 2020). This structure revealed a previously overlooked salt-bridge exhibiting a vital role in channel function and its regulation by the ‘intrinsic ligand’ motif. Therefore, using a structure-function-based approach, we strive to obtain a holistic view of the allosteric mechanisms governing KCNH channels regulation by their intracellular domains.
High-throughput identification of novel GGPPS inhibitors for multiple myeloma therapy
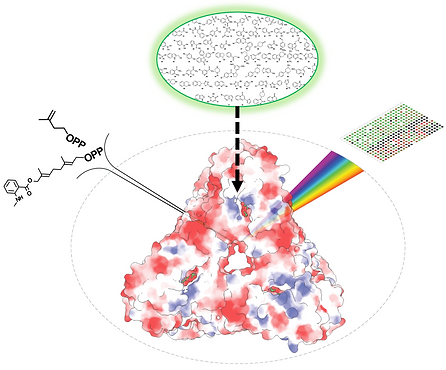
Geranylgeranyl Pyrophosphate Synthase (GGPPS) inhibition holds great potential for treating multiple myeloma (MM), a devastating condition accounting for ~1% of all cancers and ~13% of hematological malignancies. Previous efforts for developing GGPPS inhibitors focused on bisphosphonates derivatives, which have poor pharmacological profiles. Thus, in order to expand the chemical space explored, we established an innovative high-throughput screening (HTS) experimental platform, which is based on the use of a fluorescent substrate analog. Uniquely, our approach faithfully reports both on GGPPS catalysis and binding site occupancy. In this project, we employ an experimental paradigm, spanning from purified proteins to cellular analyses, to explore previously uncharted chemical space of potential GGPPS inhibitors. Our goal is T to pinpoint novel GGPPS inhibitors with translational potential in the treatment of MM.
